SysNDD: more than just a gene list! A call for NDD expert curators
– The new SysNDD database (https://sysndd.dbmr.unibe.ch) has replaced its predecessor SysID as a regularly updated, manually expert curated database on NDD associated genes and related phenotypes.
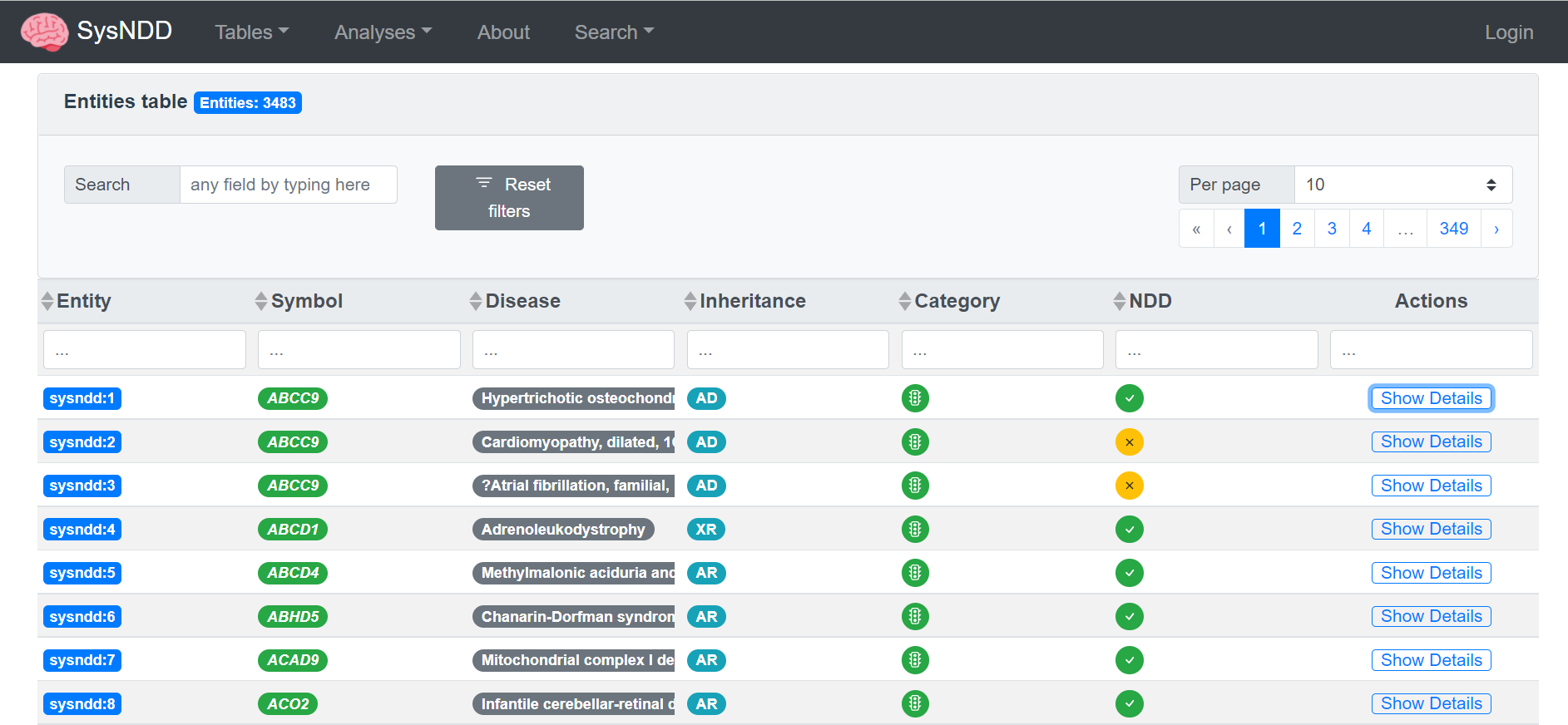
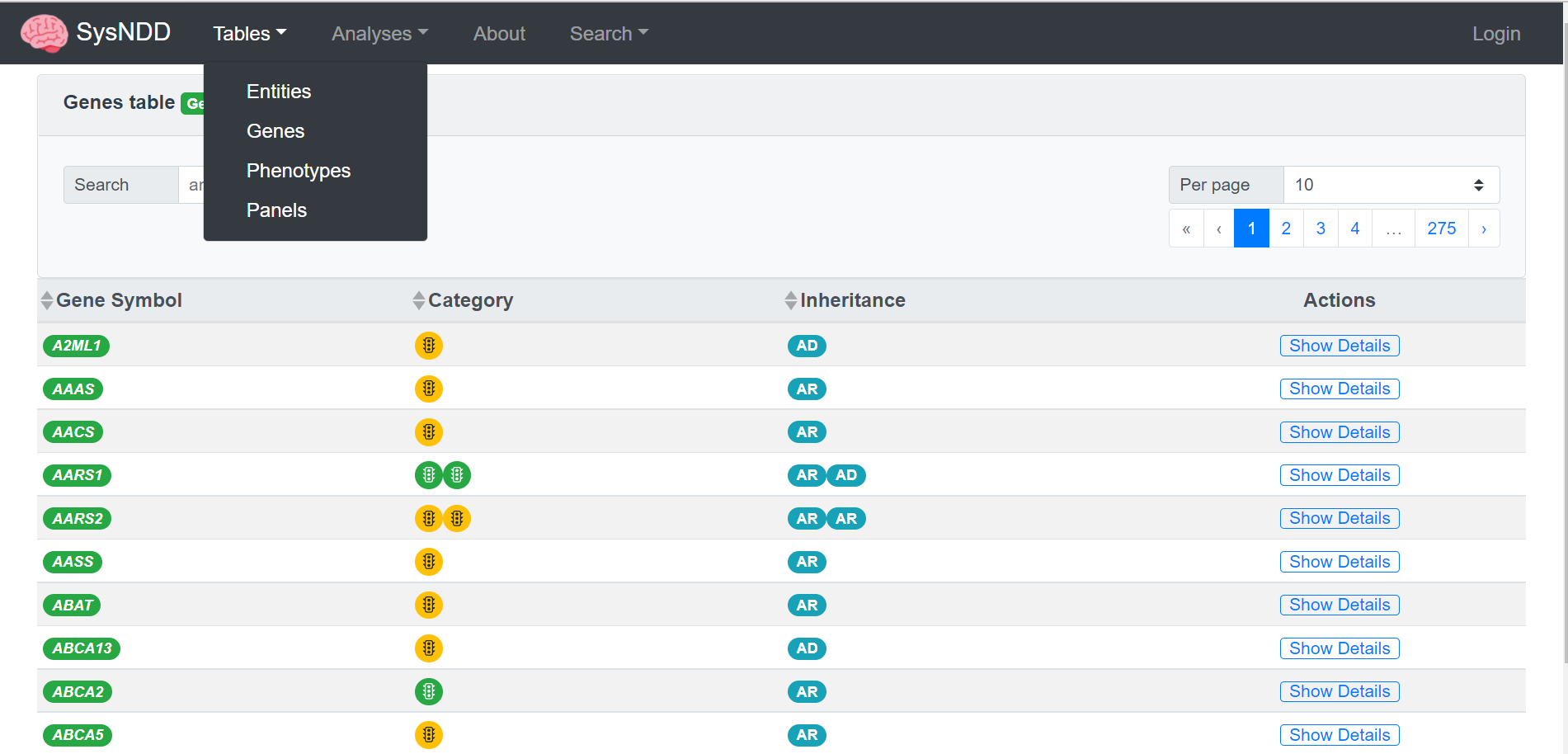
– SysNDD currently contains >1560 confirmed NDD genes and >1300 published candidate genes plus curated information on associated diseases (incl. links to OMIM, Orphanet and PubMed), phenotypes (clinical synopsis and HPO terms) and inheritance patterns. It also has manifold data comparison and filtering options, making it a valuable tool for many clinical, diagnostic and research questions. It’s Application Programming Interface (API) allows programmatic access to the data and enables sophisticated analyses and easy integration in other tools and pipelines.
– For the next months and years we will attempt a re-evaluation and curation of all entries assembled during the last 12 years. For this we are searching for expert curators.
– What am I volunteering for as an expert curator? a) Registration in the SysNDD curation tool b) Attending an online workshop (1-2 h) supplemented by online tutorials and written curation manuals. c) Each curator will receive a list of entries to curate via the curation tool. Favorite genes can be indicated and will be considered for the otherwise random assignment. d) Workload per curator (number of entries) will be determined based on a current testing phase to estimate average and min-max duration and effort per entry. d) Re-curation of the data is a one-time effort, but curators might be willing to participate in long-time curation and updating of the database.
– What are my benefits for volunteering? a) Favorite entries can be curated by experts in the field. b) Curation provides a unique opportunity to obtain an overview on the genetic and clinic landscape of NDDs and to learn about genes and diseases you might not have encountered before. c) Your work is a valuable contribution to a shared ITHACA effort for an European consensus NDD catalogue that will be closely linked to Orphanet. d) Co-authorship(s) on possible future SysNDD manuscript(s) and acknowledgement on the SysNDD website.
– If you are willing to participate or if you have questions, please contact any of the following:
The SysNDD database is based on its predecessor database, SysID (https://www.sysid.dbmr.unibe.ch/, previously: http://sysid.cmbi.umcn.nl/). It contains a manually curated catalogue of published genes implicated in neurodevelopmental disorders (NDDs) and classified into primary and candidate genes according to the degree of underlying evidence.
Furthermore, manually curated information on associated diseases and phenotypes is provided. To allow interoperability and mapping between gene-, phenotype- or disease-oriented databases, we center our approach around curated gene-inheritance-disease units, so called entities. SysNDD is updated every 3-4 months and can be utilized for a broad spectrum of tasks from both research and diagnostics.
One of our long-term goals is to incorporate the SysID / SysNDD data into other gene/disease-relationship databases like the Orphanet ontology.
SysID was initially conceived by Annette Schenck (Radboud University Center Nijmegen, the Netherlands) and Christiane Zweier (Department of Human Genetics at the University/ University Hospital Bern, Switzerland; previously: Institute for Human Genetics, University Hospital Erlangen), FAU Erlangen-Nürnberg, Germany). SysNDD has been built and is maintained by Christiane Zweier and Bernt Popp (senior physician at the Berlin Institute of Health (BIH) at Charité, Germany and scientist at the Institute of Human Genetics at the University Hospital Leipzig, Germany). CZ has been involved in establishing and updating SysID from its start in 2009. She is performing and coordinating curation and updates. BP developed and programmed the new SysNDD database and will be integrating further functionality including associated variants and network analyses in future updates.